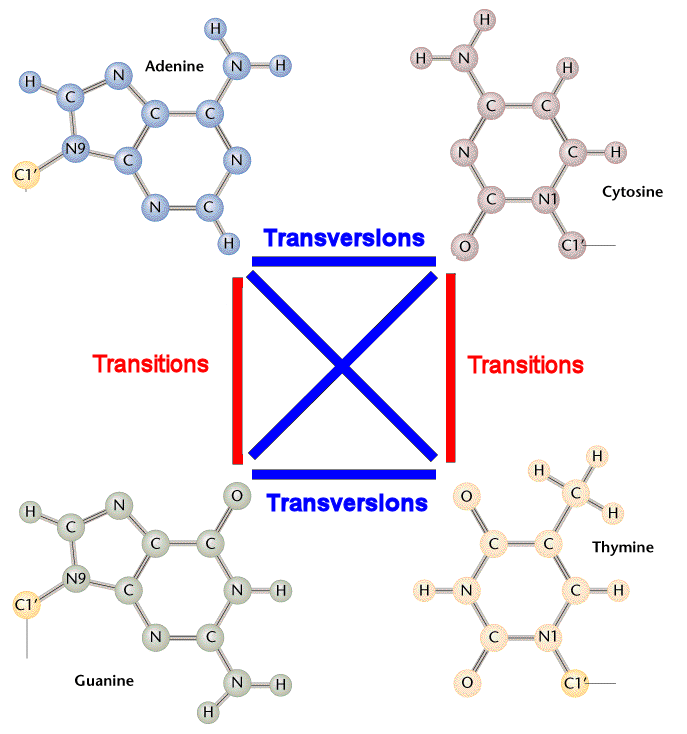
DNA substitution mutations are of two types. Transitions are
interchanges of two-ring purines
(A  G)
or of one-ring pyrimidines (C
G)
or of one-ring pyrimidines (C  T): they therefore involve
bases of similar shape. Transversions are interchanges
of purine for pyrimidine bases, which therefore involve
exchange of one-ring and
two-ring structures.
T): they therefore involve
bases of similar shape. Transversions are interchanges
of purine for pyrimidine bases, which therefore involve
exchange of one-ring and
two-ring structures.
Although there are twice as many possible transversions,
because of the molecular
mechanisms by which they are generated, transition mutations
are generated at higher frequency than transversions. As
well, transitions are less likely to result in amino acid
substitutions (due to "wobble"),
and are therefore more likely to persist as "silent
substitutions" in populations as single
nucleotide polymorphisms (SNPs).
http://www.mun.ca/biology/scarr/Transitions_vs_Transversions.html
Transition to Transversion Ratio
Human mutations don't occur randomly. In fact, transitions (changes
from A <-> G and C <-> T) are expected to occur twice as
frequently as transversions (changes from A <-> C, A <-> T, G
<-> C or G <-> T). Thus, another useful diagnostic is the
ratio of transitions to transversions in a particular set of SNP calls.
This ratio is often evaluated separately for previously discovered and
novel SNPs.
Across the entire genome the ratio of transitions to
transversions is typically around 2. In protein coding regions, this
ratio is typically higher, often a little above 3. The higher ratio
occurs because, especially when they occur in the third base of a codon,
transversions are much more likely to change the encoded amino acid. A
refinement to this analysis, in protein coding regions, is to examine
the transition to transversion ratio separately for non-degenerate,
two-fold degenerate, three-fold degenerate and four-fold degenerate
sites.
http://genome.sph.umich.edu/wiki/SNP_Call_Set_Properties
Some useful papers:
Transition-Transversion Bias Is Not Universal: A Counter Example from Grasshopper Pseudogenes
http://www.plosgenetics.org/article/info%3Adoi%2F10.1371%2Fjournal.pgen.0030022
Estimation of the transition/transversion rate bias and species sampling
http://www.ncbi.nlm.nih.gov/pubmed/10093216
Mutational and fitness landscapes of an RNA virus revealed through population sequencing
http://www.nature.com/nature/journal/v505/n7485/full/nature12861.html