cited from:
https://estrip.org/articles/read/tinypliny/44920/Linkage_Disequilibrium_Blocks_Triangles.html


https://estrip.org/articles/read/tinypliny/44920/Linkage_Disequilibrium_Blocks_Triangles.html
07/10/08 05:51 - 75ºF - ID#44920
Linkage Disequilibrium Blocks/Triangles
I
just had a zen moment in the interpretation of Linkage Disequilibrium
Maps. (Also called LD maps, LD blocks, LD triangles - take your pick.)
Turns out I was actually sweating 1st grade stuff!
I found that NO ONE explains this EXTRAORDINARILY SIMPLE thing in their umpteen papers, reviews, tutorials and what-nots. I just want to post this here so that when people google this simple little question, they find an equally simple and straight-forward answer!
This is an example of what a very small section of a Linkage Disequilibrium Map or an LD Map looks like.
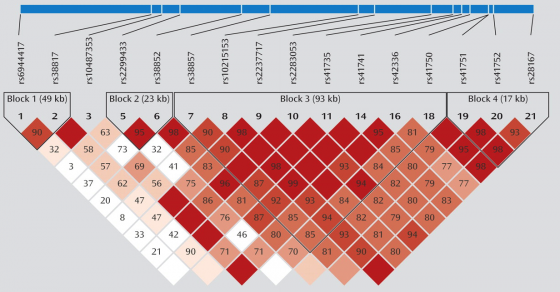
Concentrate on the upper part of the map.
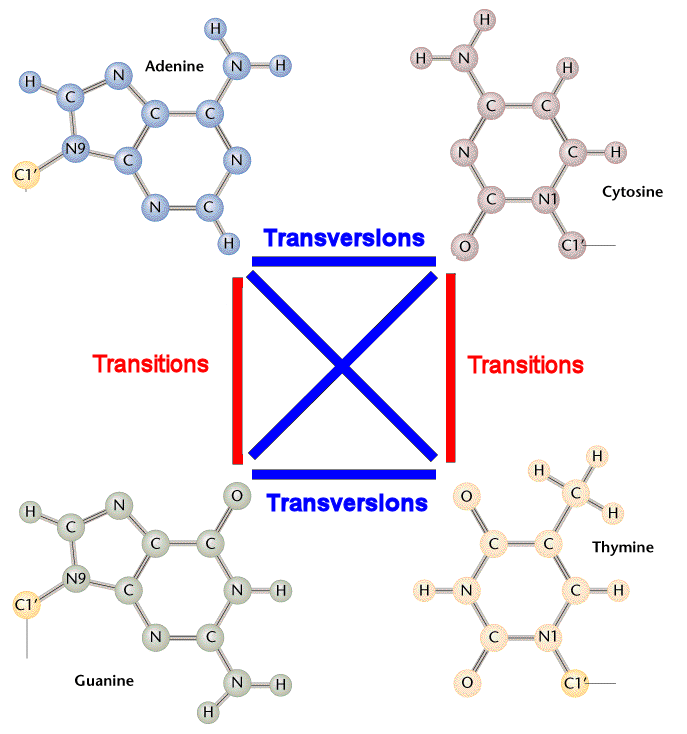
The thick blue line represents a strand of a chromosome. The white bars on the blue line of the chromosome are SNPs (Single Nucleotide Polymorphisms) that have been identified and sequenced. This means that we know what initial Nucleotide base has morphed into what final Nucleotide base. (Thus making it a polymorphic locus - or a position on the chromosome that exists in more than one form. The two forms are the intial nucleotide base and the final nucleotide base.)
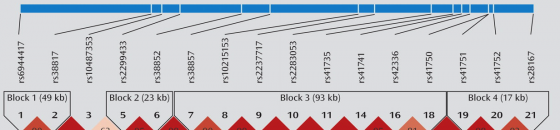
These SNP locations or loci are labeled in this picture as 1, 2, 3, ... and so on. Each of these SNPs has a name that starts with rsXXXXX where XXXXX is some numeric code. Each SNP is represented by a labeled grey triangle below the thick blue line (the chromosome).

The purpose of an LD map is to tell us whether any two given SNPs are INHERITED TOGETHER in an offspring. In other words, we want to know if any two given SNPs are in Linkage Disequilibrium.
An example: Are say, SNP #5 and SNP #9 in linkage disequilibrium? You trace down the column leading from grey triangle #5 or SNP#5 (Name: rs2299433) going toward SNP #9 (rs2237717). Do the same for SNP #9 going toward SNP #5.
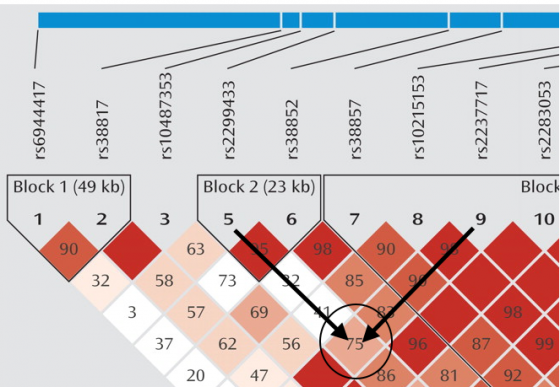
The square in which the columns leading from SNP #5 and SNP #9 intersect is the one you should focus on. I have encircled it above. As you can see its a LIGHT RED and has a number, 75. Thus SNP#5 and SNP #9 have a correlation of 0.75 and are in fairly high linkage disequilibrium with each other.
In simple terms, if your square of focus is a deep red, then the two SNPs you are interested in have the highest correlation with each other and have a highest Linkage Disequilibrium. Thus, one of them can easily act as a proxy for another. The lighter the shade of red, the lesser is the correlation between the two SNPs. For example, SNP #5 and SNP #7 have a low correlation (0.32) with each other. Thus, you cannot reliably take SNP #5 and say that it could possibly act as a proxy for SNP #7.
LD Maps also tell us about HAPLOTYPE blocks. See the blocks labeled, "Block 1 (49kb)", "Block 2 (23kb)", "Block 3 (93kb)" ... and so on.

These triangles or the blocks of dark red represent SNPs that are all in high linkage disequilibrium with each other and thus are all inherited together. They are also on the same section of the chromosome. These SNPs form a HAPLOTYPE. Every big red triangle or block in the LD map indicates a HAPLOTYPE on the corresponding stretch of the chromosome above. You only need to look at one or maximum a couple SNPs in a haplotype to know about the fate of the entire section of the chromosome that forms a Haplotype. It saves money and time.
The HapMap Consortium project has painstakingly constructed such an LD map for each and every known
SNP in the entire human genome. Their LD maps look somewhat like this
(using the haploview software:
has painstakingly constructed such an LD map for each and every known
SNP in the entire human genome. Their LD maps look somewhat like this
(using the haploview software:  )
)
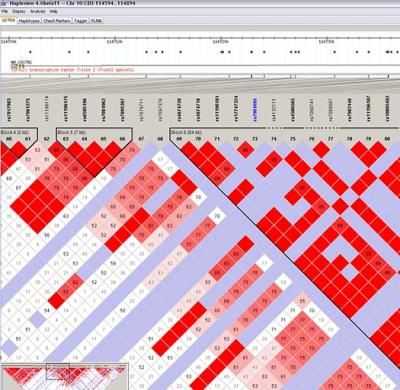
Though it is complicated, if you followed the simple tutorial above, you should be able to make sense of even complicated maps such as these. You are most welcome to leave a comment or drop me an email if you need further clarification!
I don't care who is laughing at this ridiculously detailed explanation of a kindergarten concept in genetics and genomics. Personally, I am just EXTREMELY relieved to finally know it well enough to be able to explain it. :)
I found that NO ONE explains this EXTRAORDINARILY SIMPLE thing in their umpteen papers, reviews, tutorials and what-nots. I just want to post this here so that when people google this simple little question, they find an equally simple and straight-forward answer!
This is an example of what a very small section of a Linkage Disequilibrium Map or an LD Map looks like.

Concentrate on the upper part of the map.

The thick blue line represents a strand of a chromosome. The white bars on the blue line of the chromosome are SNPs (Single Nucleotide Polymorphisms) that have been identified and sequenced. This means that we know what initial Nucleotide base has morphed into what final Nucleotide base. (Thus making it a polymorphic locus - or a position on the chromosome that exists in more than one form. The two forms are the intial nucleotide base and the final nucleotide base.)
These SNP locations or loci are labeled in this picture as 1, 2, 3, ... and so on. Each of these SNPs has a name that starts with rsXXXXX where XXXXX is some numeric code. Each SNP is represented by a labeled grey triangle below the thick blue line (the chromosome).
The purpose of an LD map is to tell us whether any two given SNPs are INHERITED TOGETHER in an offspring. In other words, we want to know if any two given SNPs are in Linkage Disequilibrium.
An example: Are say, SNP #5 and SNP #9 in linkage disequilibrium? You trace down the column leading from grey triangle #5 or SNP#5 (Name: rs2299433) going toward SNP #9 (rs2237717). Do the same for SNP #9 going toward SNP #5.

The square in which the columns leading from SNP #5 and SNP #9 intersect is the one you should focus on. I have encircled it above. As you can see its a LIGHT RED and has a number, 75. Thus SNP#5 and SNP #9 have a correlation of 0.75 and are in fairly high linkage disequilibrium with each other.
In simple terms, if your square of focus is a deep red, then the two SNPs you are interested in have the highest correlation with each other and have a highest Linkage Disequilibrium. Thus, one of them can easily act as a proxy for another. The lighter the shade of red, the lesser is the correlation between the two SNPs. For example, SNP #5 and SNP #7 have a low correlation (0.32) with each other. Thus, you cannot reliably take SNP #5 and say that it could possibly act as a proxy for SNP #7.
LD Maps also tell us about HAPLOTYPE blocks. See the blocks labeled, "Block 1 (49kb)", "Block 2 (23kb)", "Block 3 (93kb)" ... and so on.

These triangles or the blocks of dark red represent SNPs that are all in high linkage disequilibrium with each other and thus are all inherited together. They are also on the same section of the chromosome. These SNPs form a HAPLOTYPE. Every big red triangle or block in the LD map indicates a HAPLOTYPE on the corresponding stretch of the chromosome above. You only need to look at one or maximum a couple SNPs in a haplotype to know about the fate of the entire section of the chromosome that forms a Haplotype. It saves money and time.
The HapMap Consortium project
 has painstakingly constructed such an LD map for each and every known
SNP in the entire human genome. Their LD maps look somewhat like this
(using the haploview software:
has painstakingly constructed such an LD map for each and every known
SNP in the entire human genome. Their LD maps look somewhat like this
(using the haploview software:  )
)
Though it is complicated, if you followed the simple tutorial above, you should be able to make sense of even complicated maps such as these. You are most welcome to leave a comment or drop me an email if you need further clarification!
I don't care who is laughing at this ridiculously detailed explanation of a kindergarten concept in genetics and genomics. Personally, I am just EXTREMELY relieved to finally know it well enough to be able to explain it. :)
Permalink: Linkage_Disequilibrium_Blocks_Triangles.html
Words: 714
Location: Buffalo, NY
Last Modified: 08/28/14 06:01
Words: 714
Location: Buffalo, NY
Last Modified: 08/28/14 06:01